Content Types
This page contains detailed information about all content types, storable in the TOP Framework.
Table of contents
Overview
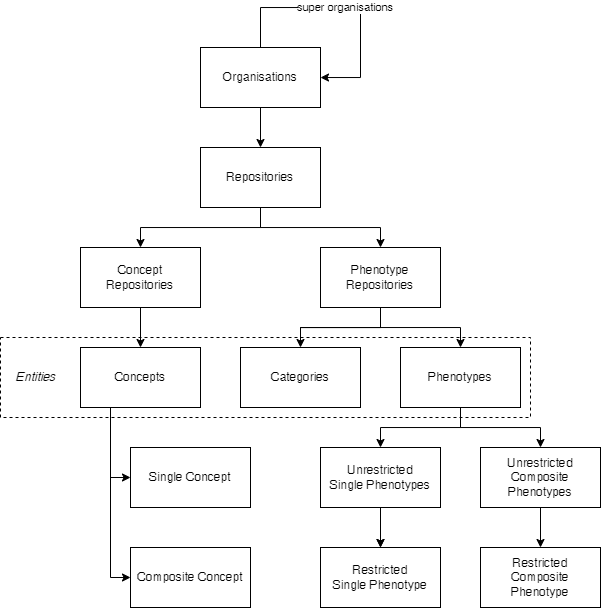
Figure 1: Hierarchy of the TOP Framework content types.
Organisations
Organisations can be used to store research groups, institutions or (scientific) projects. They can further be structured by specifying super organisations.
Repositories
Repositories are one of the main components of the TOP Framework. Depending on their type, repositories can hold different entity types. We distinguish between the following types:
Phenotype repository
In this type of repository, one can store phenotype definitions.
We recommend you to create a phenotype repository for each phenotype algorithm you want to model. By capsulising algorithms that way, you make sure that they are more comprehensible and can be found easily.
Concept repository
In contrast to the phenotype repository type, this type can only hold concepts. The purpose of this repository type is to build a collection of concepts that can be used to query for specific information, contents or documents. Consider a large indexed set of documents, one wants to search for a subset that addresses some topic.
Entities
The term ‘entity’ is a generic term to express all content types listed below.
These content types have in common that they can be referenced by URLs (deep links) and they are versioned. You can always switch between versions and restore previous ones. More details about versioning are described here.
Additionally, entities have the following properties in common:
- unique identifier
- multilingual titles, there can only be one title per language
- multilingual synonyms
- multilingual descriptions/definitions
- codes of standard terminologies, like LOINC, SNOMED CT, etc.
You can find more information about the different entity types and the underlying ontological model in our publication:
Uciteli A, Beger C, Kirsten T, Meineke FA, Herre H. Ontological Modelling and Reasoning of Phenotypes. CEUR Workshop Proceedings. 2019 Sep;2570. issn: 1613-0073. http://ceur-ws.org/Vol-2518/paper-ODLS11.pdf.
Categories
Use categories to structure repositories. They are similar to folders in a file system and serve no other purpose than organising content.
Concepts
Concepts are the main entities in concept repositories. They can be single concepts or composite concepts. Single concepts are sets of synonyms that are used to represent a specific idea or topic. Composite concepts are constructed from one or more single or composite concepts and can be used to express more complex ideas or topics. They are defined using logical operators such as AND, OR, NOT, and more advanced operators.
Phenotypes
Phenotypes are another one of the main components of the TOP Framework. We use the following definition for the term ‘phenotype’:
“A (combination of) bodily feature(s) of an organism determined by the interaction of its genetic make-up and environment.”
Scheuermann RH, Ceusters W, Smith B. Toward an ontological treatment of disease and diagnosis. Summit Transl Bioinform. 2009 Mar 1;2009:116-20. PMID: 21347182; PMCID: PMC3041577.
The phenotypes you model in the TOP Framework are actually classes. The phenotypes of individuals (organisms, people, patients, …) are members of these classes. For example, the height of the person John Doe is a member of the phenotype class ‘Height’ and has the value 180 centimetres.
All phenotypes have the mandatory property ‘data type’, which can be one of numeric, date time, text and Boolean. Numeric phenotypes may also have a unit of measurement, specified as Unified Code for Units of Measure (UCUM).
We distinguish between single and composite phenotypes, which are described below.
Single Phenotypes
Basic properties of individuals are modelled as single phenotypes. They can also be called ‘atomic’ or ‘elemental’, because of their indivisible nature. All single phenotypes should have at least one code associated with them, so that they can be mapped to individual data in any kind of data holding source system.
Composite Phenotypes
In contrast to single phenotypes, composite ones are constructed from one or more single or composite phenotype or constant (arguments). This is done by applying functions (e.g., addition, subtraction, logical ‘and’ operation, etc.) that transform a set of input arguments to typically one value.
Example:
The expression of the phenotype ‘BMI’ (body mass index) could look like:
Weight [kg] / (Height [cm] / 100) ^ 2
Difference Between Unrestricted and Restricted Phenotypes
Single and composite phenotypes can further be divided in unrestricted and restricted phenotypes. Unrestricted phenotypes represent all possible phenotypes of individuals, whereas restricted phenotypes have further restrictions that must be matched in order for individual phenotypes to be classified to them.
Example:
All possible heights of individuals are classified to the unrestricted phenotype ‘Height’ and only a portion of them are classified to the restricted phenotype ‘Height > 200 cm’.
Phenotype Algorithms
Phenotype algorithms are depicted by a set of single and/or composite phenotypes. Typically there is a composite phenotype that references all other phenotypes in it’s expression. To execute a phenotype algorithm, one has to provide data of an individual and evaluate all expressions and restrictions in the set.
There is an extensive example for an algorithm to detect Diabetes Mellitus Type 2 available in:
Uciteli A, Beger C, Kirsten T, Meineke FA, Herre H. Ontological representation, classification and data-driven computing of phenotypes. J Biomed Semantics. 2020 Dec 21;11(1):15. doi: 10.1186/s13326-020-00230-0.